
Breeding theory
Multi-environment trials – design and analysis
.gif)
To clarify the purpose of variety trials
To introduce linear models for multi-environment trials
To describe the analysis of variance for METs
To model the variance of a cultivar mean estimated from a MET
To examine the effect of replication within and across sites and years on measures of precision.
To use standard error and LSD modeling to compare different allocations of testing resources for predictive power, cost, and efficiency.
Introduction
As observed in previous units, the precision and predictive power of individual field trials is very low. The most important tool for predicting cultivar performance within the TPE is the multi-environment trial (MET). New cultivars are tested at several locations and over several years at locations that sample the TPE. The true values of cultivars are unknown. Cultivar means estimated from METs are the best predictors of future cultivar performance, but they are always estimated with error. This error can be minimized by increasing the number of replicates, sites, and years of field testing, but conducting METs is very expensive, requiring much of the time and money available to plant breeding programs.
It is important that testing programs be designed to use these resources efficiently, and to maximize the precision of estimation of cultivar means. In this unit, the design and analysis of METs will be presented using software that is available to NARES researchers. Tools for assessing the precision of METs will be presented. You will learn how to decide how to allocate testing effort (sites, years, and replications) to maximize the precision and predictive power a variety testing program, given the personnel, funding, and time available to you.
1. The purpose of variety trials
The purpose of a variety trial is to predict the performance of new varieties, relative to a check, in farmers’ fields and in future seasons within the TPE
It is very important to understand that the real purpose of a variety trial in a breeding program is prediction. The field and season in which the trial is conducted is considered to be a random sample of farmers’ fields and future seasons in the TPE. The recognition that the individual trial environment is a random factor has important implications for our understanding of the precision of a trial, or its power to detect differences in genotypic value among lines in the trial can be observed. The precision of a variety trial is analogous to the magnifying power of a microscope; a high level of precision in a variety trial is needed to detect a small difference in the genotypic value of breeding lines. A lower level of precision is needed to detect a large difference between varieties. The precision of a variety trial is mainly determined by its level of replication within and across environments. The relative precision of different variety trials can be compared by their SEM or LSD.
2. Linear models describing measurements obtained from field trials
The genotype x environment (GE) model
We conduct the combined analysis of variety trials over locations and years within the TPE to estimate the mean performance of varieties. Estimates of the precision of the means (SEM and LSD) are also required. To estimate the variance of a cultivar mean, we must use a statistical model that describes the factors or sources contributing to that variance. The simplest GEI model for the analysis of a MET is:
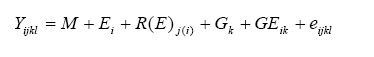
where:
|
Yijkl |
= the measurement on plot l in environment i, block j, containing genotype k |
|
M |
= the overall mean of all plots in all environments |
|
Ei |
= the effect of environment (trial) i |
|
R(E)j(i) |
= the effect of replicate j within environment i |
|
Gk |
= the effect of genotype k |
|
GEik |
= the interaction of genotype i with environment k |
|
eijkl |
= the plot residual |
In this model, genotype effects are usually considered fixed; that is, we wish to estimate the performance of the specific genotypes in the trial. Environments and replicates are random factors; we are not interested in the means of the individual trials per se, but rather are interested in them only as sampling the TPE. (Occasionally, genotypes may also be considered random, if the purpose of the trial is to estimate genetic variances rather than to predict cultivar performance.)
GE interactions are random in this model, because the interaction between fixed and random factors is always random. The random GE term contributes to the true error in the test of differences among cultivars, and to the variance of cultivar means.
According to this model, the variance of a cultivar mean is:
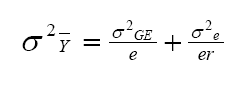
where 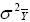 is the variance of a cultivar mean, e is the number of trials,
and r is the number of replicates per trial.
is the variance of a cultivar mean, e is the number of trials,
and r is the number of replicates per trial.
It is important to minimize 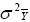 in cultivar testing programs.
in cultivar testing programs.
Minimizing 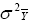 leads
to improved prediction of cultivar performance in the future, and increased
gains from breeding.
leads
to improved prediction of cultivar performance in the future, and increased
gains from breeding.
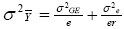 can be used to determine the minimum value for
can be used to determine the minimum value for 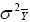 that
can be achieved with the resources available to the breeder.
that
can be achieved with the resources available to the breeder.
The variance components can be estimated from the ANOVA table for a completely balanced set of trials (all varieties tested in all trials). Methods for estimating variance components from unbalanced data sets are available but require more sophisticated software.
The expected mean squares from the ANOVA of a MET are linear functions
of the variances of the factors in 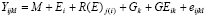 .
.
These are given below:
|
Source |
Mean square |
EMS |
|
Environments (E) |
|
|
|
Replicates within E |
|
|
|
Genotypes (G) |
MSG |
σ2e+ rσ2GE + rgσ2G |
|
G x E |
MSGE |
σ2e + rσ2GE |
|
Plot residuals |
MSe |
σ2e |
Table: Expected mean squares (EMS) for the ANOVA of the genotype x environment model assuming all factors random
The variance components can be estimated as functions of the mean squares estimated from the ANOVA:
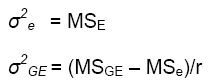
It should be noted that these variance components have very large standard errors, and should be used only as a rough guide to planning a breeding or testing program. They should only be estimated if the number of degrees of freedom for the mean square is about 50 or greater.
Example 1: Resource allocation for a rice breeding program using the GE model (click on the icon)
Example 2: Resource allocation for a rice breeding program using the GE model (click on the icon)
Deciding whether to divide a target population of environments into two regions for breeding purposes
The analyses above are useful tools for deciding how many locations, years and replicates of testing are needed to predict performance of a variety with adequate precision. They do not, however, give guidance about whether the TPE should be broken into 2 breeding targets.
There are many genotype x environment interaction analyses that can be used to group sites into relatively similar groups based on cultivar performance. These methods, including cluster, pattern, and AMMI analyses, are useful when there is no pre-existing hypothesis to test about the pattern of adaptation of varieties to sites.
However, when there is a good hypothesis to test (based on breeder knowledge, farmer practice, or agroclimatic data), there is a straightforward model that can be used for testing whether locations within the TPE can really be grouped into more homogeneous subsets. In this model, trial sites are grouped into subgroups based on location or some other fixed factor.
For example, if a breeder wished to evaluate whether two subregions, say the northern and southern parts of the TPE, should be considered separate breeding targets, he or she would classify all trials in the north into one group, and all fields in the south into a second group. A combined analysis would be performed over trials, with the location factor broken down into two subcomponents: subregion (north or south) and trials within subregion.
This is illustrated for the 2-way GxE model
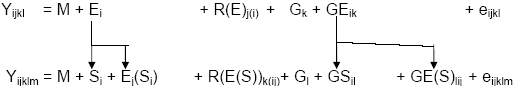
In this model, subregions are considered fixed effects. Locations within subregions are a random sampling factor, like replicates or years. The hypothesis that varieties perform very differently in different regions can be tested by testing the variety x subregion mean square against the variety x locations within subregion mean square. (This model can be easily extended to the genotype x location x year model). Because this is a mixed model the analysis needs to be done with software that can support mixed model analyses, including the latest release of IRRISTAT.
The ANOVA table with expected mean squares for a balanced case is given below:
Table 5. Expected mean squares (EMS) for the ANOVA of the genotype x subregion for testing fixed groupings of trial sites
|
Source |
Mean square |
EMS |
|
Subregions (S) |
|
|
|
Locations within subregions (L(S)) |
|
|
|
Replicates within L(S) |
|
|
|
Genotypes (G) |
MSG |
σ2e + rσ2GL(S) + rlσ2GS + rlsσ2G |
|
G x S |
MSGS |
σ2e + rσ2GL(S) + rlσ2GS |
|
G x L(S) |
MSGL(S) |
σ2e + rσ2GL(S) |
|
Plot residuals |
MSe |
σ2e |
Example of a test of significance of genotype x subregion interaction
Breeders in Laos would like to know if the southern and central parts of the country need to be considered separate TPE. A trial involving 22 tradition varieties was conducted at 6 locations in the wet season of 2004, three in the center and three in the south.
The ANOVA table is presented below:
Table: ANOVA for 22 varieties tested in two Lao PDR subregions (north and south), with 3 locations per subregion, in Wet season 2004
|
Source |
df |
MS |
F |
|
Subregions (S) |
1 |
5459785 |
|
|
Locations within subregions (L(S)) |
4 |
17284169 |
|
|
Replicates within L(S) |
18 |
292059 |
|
|
Genotypes (G) |
21 |
3644949 |
4.77** |
|
G x S |
21 |
764412 |
0.76 |
|
G x L(S) |
84 |
1006974 |
6.58** |
|
Plot residuals |
378 |
153101 |
|
In this case, the genotype x subregion (G x S) term is not significant when tested against the pooled genotype x location within subregion (G x L(S)) term.
Division of this TPE into 2 subregions base on central versus southern location therefore is unwarranted, at least on the basis of this 2004 trial. A combined analysis of an experiment conducted over several years would give a more reliable result.
Let's conclude
Summary
The purpose of a variety trial is to predict future performance in the TPE
Genotype x environment interaction is large, and reduces the precision with which cultivar means can be estimated.
Variance component estimates for the GLY model can be used to develop testing programs that maximize precision for a given level of resources.
Within relatively homogeneous TPE, the genotype x site x year variance component is usually the largest. When this is the case, strategies that emphasize either testing over several sites or testing over several years are likely to be successful.
Little benefit is obtained from including more than 3 replicates (and often more than 2) in a MET.
Standard errors and LSDs estimated from single sites are unrealistically low because they do not take into account genotype x environment interaction
A decision about whether a TPE should be split into 2 separate breeding targets can be made by grouping trial sites or locations into subgroupings based on some fixed environmental factor, and then testing the significance of the genotype x subgroup interaction.
References
Cooper, M., Rajatasereekul, S., Immark, S., Fukai, S., Basnayake, J. 1999b. Rainfed lowland rice breeding strategies for northeast Thailand. I. Genotypic variation and genotype environment interactions for grain yield. Field Crops Research 64: 131-151.
Atlin, G.N, R.J. Baker, K.B. McRae, and X. Lu. 2000. Selection response in subdivided target regions. Crop Sci. 40:7-13.
Next lesson
In the next lesson, we will discuss broad sense heritability estimates and selection response.
