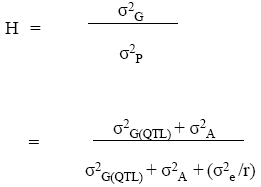|
|
|
|

| ||
|
|
|
Molecular breeding |
|||
|
|
|
Quantitative trait locus (QTL) mapping |
|||
|
|
|
|
|||
|
|
|
|
|||
|
|
|
1. What is QTL?
QTL = a gene or chromosomal region that affects a quantitative trait.
It must be polymorphic (have allelic variation) to have an effect in a population and it must be linked to a polymorphic marker allele to be detected
|
| ||
|
|
|
2. Mapping quantitative trait loci (QTL)
QTL = underlying genes controlling quantitative traits
|
| ||
|
|
|
Example: In progeny derived from cross AA x aa:
Mean of AA lines is 3100 ± s.e.m Mean of aa lines is 2900 ± s.e.m
BUT, AA and aa individuals can’t be visually distinguished
--> Some AA lines will have low yield due to e’s or other genes
--> Some aa lines will have high yield due to e’s or other genes
|
| ||
|
|
|
3. The QTL effect
|
| ||
|
|
|
4. Single-marker analysis
DNA markers can be used to map useful genes using recombination frequencies of linked genes:
|
| ||
|
|
|
5. Effect of a marker linked to a QTL
Recombination between M and A is R
In RILs derived from MmAa F1, individuals with MM marker genotype are made up of 2 QTL genotypes: AA & aa
-->So, the effect of marker M is a function of:
MM lines are easily distinguished from mm lines, but AA lines can’t be distinguished from aa lines. If M and A are linked, average of MM lines will differ from the average of mm lines. The size of difference can be between 0 and a, depending on the marker-QTL distance
Means of MM and mm recombinant inbred lines
|
| ||
|
|
|
6. QTL mapping with molecular markers
DNA markers used to map useful genes using recombination frequencies of linked genes:
|
| ||
|
|
|
7. QTL mapping strategies
All marker-based mapping experiments have same basic strategy:
|
| ||
|
|
|
7.1 QTL mapping strategies: Single-marker analysis
|
| ||
|
|
|
|
| ||
|
|
|
example: 25 RILs produced from an F1 between 2 homzygous parents Parents differ at marker loci A, B, and C on 1 chromosome:
Lines are evaluated in 4-rep trial
--> Is there a QTL in this region?
|
| ||
|
|
|
Measure of QTL contribution to σP2
Recall that the simplest QTL model divides the genotypic effect into a QTL effect (A) and an effect of all other genes within QTL classes (G(QTL)):
|
| ||
|
|
|
Measure of marker contribution to σP2
|
| ||
|
|
|
F-test for the difference between marker genotype classes is highly significant at locus B
Therefore, there is a QTL at or near marker B |
| ||
|
|
|
Broad-sense heritability for a trial in which 1 QTL is detected
|
| ||
|
|
|
R2 is the proportion of σP2 explained by the QTL A
|
| ||
|
|
|
Problems with single-marker analysis:
|
| ||
|
|
|
7.2 QTL mapping strategy: Interval mapping
|
| ||
|
|
|
Finding the position of QTL with molecular markers
DNA markers can be used to map useful genes using recombination frequencies of linked genes:
Recombinant gametes: M1a, m1A, Parental gametes: M1A, m1a, Frequency of recombinants is map distance
|
| ||
|
|
|
What are the problems with interval mapping?
Can’t resolve 2 QTL in a marker interval Although the LOD thresholds seem very high, too many QTLs are declared (all methods do) Ignores epitasis Not accurate for QTL with small effects (no methods are)
|
| ||
|
|
|
Linkage mapping with molecular markers
Double crossover products look like parental types, leading to map distance underestimates:
Haldane and Kosambi mapping functions used to correct recombination frequencies |
| ||
|
|
|
Significance test: Logarithm of the odds ratio (LOD score):
|
| ||
|
|
|
8. Fine mapping
|
| ||
|
|
|
9. Marker-assisted backcrossing
|
| ||
|
|
|
10. How is QTL mapping best used?
|
| ||
|
|
|
11. Some guidelines for successful QTL mapping
|
| ||
|
|
|
12. Using QTL in breeding
e.g. disease resistance genes, Sub1
|
| ||
Let's conclude |
|
Summary
|
| ||
|
|
|
References:
Kearsey, M.J. and Pooni, H.S. 1996. The genetical analysis of quantitative traits. Chapter 7
Bernardo, R. 2002. Breeding for quantitative traits in plants. Chapters 13 and 14
|
| ||
Next lesson |
|
This ends module 5. The 6th and last module is about participatory approaches. This information will be very valuable for breeders when communicating and working with farmers. |
| ||
|
|
|
|

|

.gif)